 | | PATHWAY: map00400 | |
| Entry |
|
|---|
| Name |
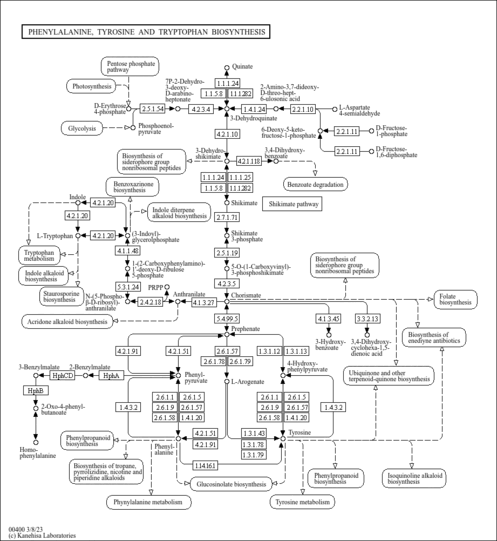
Phenylalanine, tyrosine and tryptophan biosynthesis |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
| map00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
|
|---|
| Module |
| M00022 | Shikimate pathway, phosphoenolpyruvate + erythrose-4P => chorismate [PATH:map00400] |
| M00023 | Tryptophan biosynthesis, chorismate => tryptophan [PATH:map00400] |
| M00024 | Phenylalanine biosynthesis, chorismate => phenylpyruvate => phenylalanine [PATH:map00400] |
| M00025 | Tyrosine biosynthesis, chorismate => HPP => tyrosine [PATH:map00400] |
| M00040 | Tyrosine biosynthesis, chorismate => arogenate => tyrosine [PATH:map00400] |
| M00910 | Phenylalanine biosynthesis, chorismate => arogenate => phenylalanine [PATH:map00400] |
|
|---|
| Other DBs |
|
|---|
Related
pathway |
| map00130 | Ubiquinone and other terpenoid-quinone biosynthesis |
| map00403 | Indole diterpene alkaloid biosynthesis |
| map00710 | Carbon fixation in photosynthetic organisms |
| map00950 | Isoquinoline alkaloid biosynthesis |
| map00960 | Tropane, piperidine and pyridine alkaloid biosynthesis |
| map01053 | Biosynthesis of siderophore group nonribosomal peptides |
| map01059 | Biosynthesis of enediyne antibiotics |
|
|---|
| KO pathway |
|
|---|
|
|
|