KEGG Pathway Map
| Pathway map |
The KEGG PATHWAY database is a collection of manually drawn graphical diagrams, called KEGG pathway maps, for metabolic pathways, signaling pathways, pathways involved in various cellular processes and organismal systems, and perturbed pathways associated with human diseases.
The pathway map identifier takes the form of:
<prefix><five-digit number>
where the number is uniquely assigned to each pathway map and the prefix is for a different version of the same pathway map, such as "map" for the manually drawn reference pathway and the three- or four-letter organism code for a computationally generated organism specific pathway.
The "Pathway menu" at the top is for changing the five-digit map number and the "Change pathway type" button is for changing the prefix.
|
|---|---|
| Side panel |
The pathway map viewer comes with a side panel for various client-side operations shown below. Option
Scale slider in all maps
Search
Module link mode in global (01100s) and overview (01200s) metabolic pathways Coloring yes or no in organism-specific global maps
Search box for searching map objects by IDs or aliase
ID search
KEGG identifier search with OR operation for multiple IDs
Color
Plus sign allows input of user data in the form of KEGG Mapper Search tool
Plus sign allows input of user data in the form of KEGG Mapper Color tool
Module
Enter KEGG identifiers followed by color specification (see details here) User data automatically stored in the local storage of the web browser
Module list in selected metabolic pathways
Reaction module
Completeness may be selected in organism-specific pathways
Reaction module list in overview (01200s) metabolic pathways
Network
Network list in selected reference and human pathways linking to network variation maps
Related Brite / Related Brite table
Links to related Brite hierarchies and Brite tables
The side panel may be collapsed/expanded and its width may be changed. |
| Short cut key | Short cut key Ctrl+F (Cmd+F or Alt+F) may be used to focus on the Search box |
|
Special map Notation | The global maps (map numbers 01100s) and the overview maps (map numbers 01200s) are a special class of metabolic pathway maps. They are drawn with lines (or arrows) as the main objects for linking KOs, enzymes, reactions and individual genes, rather than boxes in regular metabolic pathway maps. In addition, the reference global maps are drawn with coloring to distinguish eleven functional categories 09101 to 09111 in KEGG Color Codes. |
|
Regular map Notation |
All the regular maps are drawn with the notation shown below.
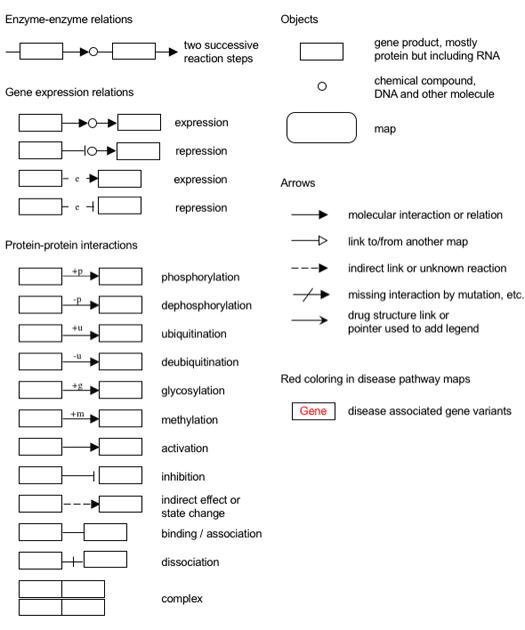
|
|
Color coding |
The pathway map without coloring is the original version that is manually drawn by in-house software called KegSketch. The other pathway maps with coloring are all computationally generated as summarized below.
|
| Image file | Use the "Download" link to obtain an image file of the pathway map as it appears on the screen (with coloring, etc.), optionally as a double (2x) sized image. In order to generate a publication-quality image, convert the resolution from 72 dpi to around 350 dpi using a software tool such as Photoshop or an online tool. |