DiGAlign
: the Dynamic Genomic Alignment server
version 2.0
DiGAlign
: the Dynamic Genomic Alignment server
version 2.0
About
DiGAlign is a genome alignment viewer that extends the alignment visualizing function implemented in ViPTree.
Nucleotide- (BLASTn) or amino acid- (tBLASTx) resolved alignments of input nucleotide sequences are computed and interactively visualized to generate a publication-ready figure.
Same as ViPTree, the alignment view can include gene and function predictions performed by the DiGAlign server or provided by the user.
DiGAlign quickly and effectively assists you in visualizing the results as you want in comparative genomics and genome structure exploration.
Workflow
Generate alignments of your query sequences.
- Upload your nucleotide sequences and choose the alignment and gene prediction options.
- Browse the alignments
- Browse dotplots / genomic alignments between your query sequences
- Download figures and tables
Example
Here is an exmaple of the session main page (an entrance page after the computation).
Snapshots

Reordering and position optimization
Previous
Next
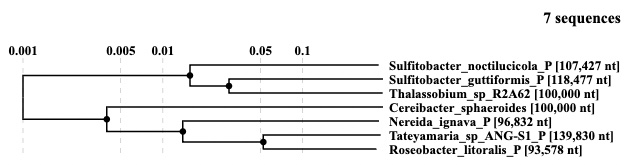
- The alignment view: An automaticaly position adjusted alignment of four anoxygenic photosynthetic organisms of the class Alphaproteobacteria were used to characterize regions containing photosynthetic gene clusters (PGCs).
- The tree view: A "guide tree" including flexible selector for picking up closely related sequences (a dot in the tree is a link for the alignment including sequences under the dot).
- A control panel of various tuning parameters for the alignment view.
- A control panel of reordering, postion optimization, duplication and deletion of the sequences.
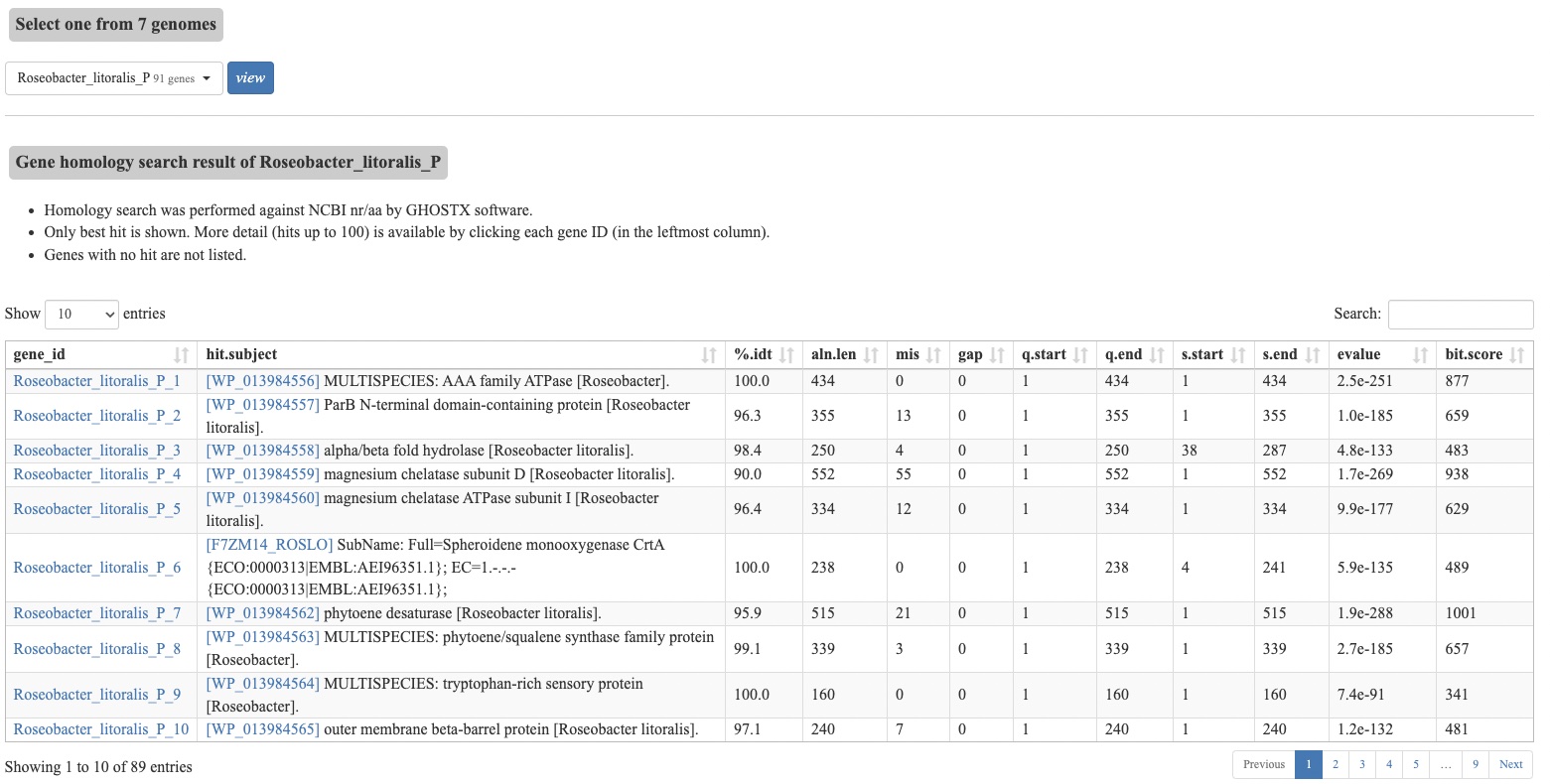
- A table of similarity search results, using the GenomeNet nr-aa database.
Version history
- 2023-11-17 version 2.0
- Major update! Various functions are newly implemented, as listed below.
- Gene prediction is now performed by Prodigal and any coding table is selectable if it is compatible with Prodigal.
- In the alignment viewer, threshold for tBLASTx/BLASTn could be set.
- In the alignment viewer, a mouseover popup is implemented that shows details of genes and tBLASTx/BLASTn hits.
- A mouseover popup is implemented that shows details of genes and tBLASTx/BLASTn hits.
- From this version, License is set as 'CC BY-NC-SA 4.0'
- Many bug fixes, including the gene table upload function.
- Document is updated.
- 2022-04-18 version 1.0
- Initial release of DiGAlign
Citation
If you use results (data / figures) generated by DiGAlign in your research, please cite:
- Nishimura Y, Yamada K, Okazaki Y, Ogata H. (2024) DiGAlign: versatile and interactive visualization of sequence alignment for comparative genomics. Microbes and Environments, 39(1), ME23061. doi: 10.1264/jsme2.ME23061 [Open Access]
Feedback
We appreciate bug reports, comments, and suggestions. The form is
here.